2016-2017 Influenza Season Week 1 ending January 7, 2017: Influenza activity increased in the United States.
January 14th, 20172016-2017 Influenza Season Week 1 ending January 7, 2017
All data are preliminary and may change as more reports are received.
Synopsis:
During week 1 (January 1-7, 2017), influenza activity increased in the United States.
- Viral Surveillance: The most frequently identified influenza virus subtype reported by public health laboratories during week 1 was influenza A (H3). The percentage of respiratory specimens testing positive for influenza in clinical laboratories increased.
- Pneumonia and Influenza Mortality: The proportion of deaths attributed to pneumonia and influenza (P&I) was below the system-specific epidemic threshold in the National Center for Health Statistics (NCHS) Mortality Surveillance System.
- Influenza-associated Pediatric Deaths: Three influenza-associated pediatric deaths were reported.
- Influenza-associated Hospitalizations: A cumulative rate for the season of 7.1 laboratory-confirmed influenza-associated hospitalizations per 100,000 population was reported.
- Outpatient Illness Surveillance: The proportion of outpatient visits for influenza-like illness (ILI) was 3.2%, which is above the national baseline of 2.2%. All 10 regions reported ILI at or above their region-specific baseline levels. New York City, Puerto Rico, and eight states experienced high ILI activity; six states experienced moderate ILI activity; seven states experienced low ILI activity; 28 states experienced minimal ILI activity, and the District of Columbia and one state had insufficient data.
- Geographic Spread of Influenza: The geographic spread of influenza in Puerto Rico and 21 states was reported as widespread; Guam and 21 states reported regional activity; the District of Columbia and eight states reported local activity; and the U.S. Virgin Islands reported no activity.
National and Regional Summary of Select Surveillance Components
| HHS Surveillance Regions* | Data for current week | Data cumulative since October 2, 2016 (week 40) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Out-patient ILI† | Number of jurisdictions reporting regional or widespread activity§ | % respiratory specimens positive for flu in clinical laboratories‡ | A(H1N1)pdm09 | A (H3) | A (Subtyping not Performed) <!–(Subtyping not performed)–> |
B Victoria lineage | B Yamagata lineage | B lineage not performed | Pediatric Deaths | |
| Influenza test results from public health laboratories only | ||||||||||
| Nation | Elevated | 44 of 54 | 13.9% | 177 | 5,261 | 134 | 174 | 123 | 147 | 3 |
| Region 1 | Elevated | 5 of 6 | 11.6% | 15 | 266 | 0 | 0 | 3 | 5 | 0 |
| Region 2 | Elevated | 3 of 4 | 15.5% | 1 | 285 | 8 | 16 | 8 | 20 | 0 |
| Region 3 | Elevated | 4 of 6 | 9.6% | 17 | 651 | 10 | 21 | 25 | 14 | 0 |
| Region 4 | Elevated | 6 of 8 | 12.1% | 15 | 355 | 16 | 17 | 12 | 66 | 1 |
| Region 5 | Elevated | 5 of 6 | 7.2% | 17 | 659 | 13 | 60 | 23 | 15 | 0 |
| Region 6 | Elevated | 4 of 5 | 8.0% | 21 | 124 | 0 | 11 | 8 | 8 | 1 |
| Region 7 | Elevated | 2 of 4 | 7.9% | 5 | 204 | 10 | 12 | 13 | 4 | 1 |
| Region 8 | Elevated | 6 of 6 | 16.8% | 32 | 608 | 8 | 9 | 11 | 0 | 0 |
| Region 9 | Elevated | 5 of 5 | 10.4% | 52 | 1,522 | 64 | 21 | 20 | 8 | 0 |
| Region 10 | Elevated | 4 of 4 | 29.8% | 2 | 587 | 5 | 7 | 0 | 7 | 0 |
*HHS regions (Region 1 CT, ME, MA, NH, RI, VT; Region 2: NJ, NY, Puerto Rico, US Virgin Islands; Region 3: DE, DC, MD, PA, VA, WV; Region 4: AL, FL, GA, KY, MS, NC, SC, TN; Region 5: IL, IN, MI, MN, OH, WI; Region 6: AR, LA, NM, OK, TX; Region 7: IA, KS, MO, NE; Region 8: CO, MT, ND, SD, UT, WY; Region 9: AZ, CA, Guam, HI, NV; and Region 10: AK, ID, OR, WA).
† Elevated means the % of visits for ILI is at or above the national or region-specific baseline
§ Includes all 50 states, the District of Columbia, Guam, Puerto Rico, and U.S. Virgin Islands
‡ National data are for current week; regional data are for the most recent three weeks
U.S. Virologic Surveillance:
WHO and NREVSS collaborating laboratories, which include both public health and clinical laboratories located in all 50 states, Puerto Rico, and the District of Columbia, report to CDC the total number of respiratory specimens tested for influenza and the number positive for influenza by virus type. In addition, public health laboratories also report the influenza A subtype (H1 or H3) and influenza B lineage information for the viruses they test and the age or age group of the persons from whom the specimens were collected.
Additional data are available at http://gis.cdc.gov/grasp/fluview/fluportaldashboard.html and http://gis.cdc.gov/grasp/fluview/flu_by_age_virus.html.
The results of tests performed by clinical laboratories during the current week are summarized below.
| Week 1 | Data Cumulative since October 2, 2016 (Week 40) |
|
|---|---|---|
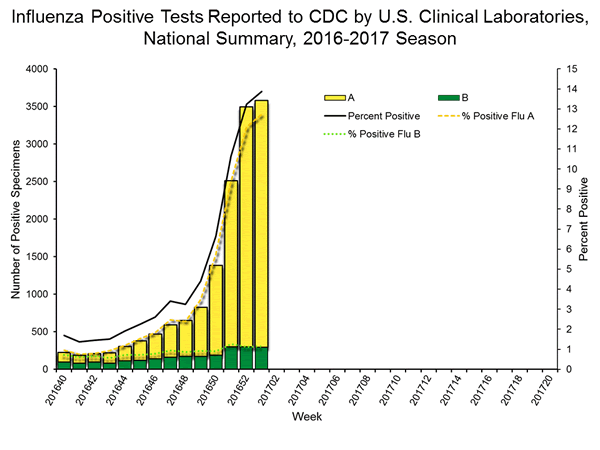
| No. of specimens tested | 25,797 | 259,647 |
| No. of positive specimens (%) | 3,580 (13.9%) | 15,026 (5.8%) |
| Positive specimens by type | ||
| Influenza A | 3,287 (91.8%) | 12,709 (84.6%) |
| Influenza B | 293 (8.2%) | 2,317 (15.4%) |
The results of tests performed by public health laboratories, as well as the age group distribution of influenza positive tests, during the current week are summarized below.
| Week 1 | Data Cumulative since October 2, 2016 (Week 40) |
|
|---|---|---|
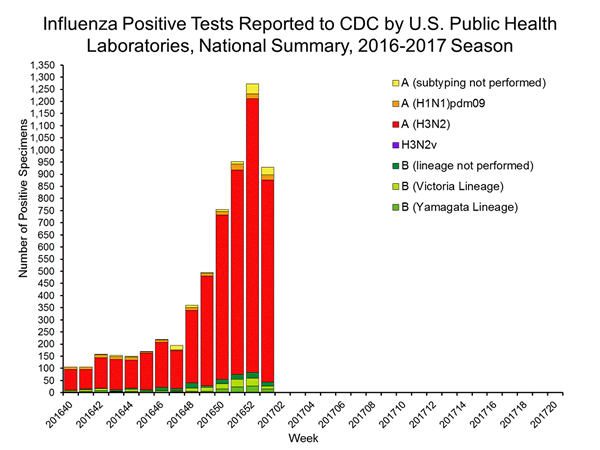
| No. of specimens tested | 1,973 | 21,365 |
| No. of positive specimens* | 928 | 6,016 |
| Positive specimens by type/subtype | ||
| Influenza A | 885 (95.4%) | 5,572 (92.6%) |
| A(H1N1)pmd09 | 21 (2.4%) | 177 (3.2%) |
| H3 | 833 (94.1%) | 5,261 (94.4%) |
| Subtyping not performed | 31 (3.5%) | 134 (2.4%) |
| Influenza B | 43 (4.6%) | 444 (7.4%) |
| Yamagata lineage | 15 (34.9%) | 123 (27.7%) |
| Victoria lineage | 12 (27.9%) | 174 (39.2%) |
| Lineage not performed | 16 (37.2%) | 147 (33.1%) |
*The percent of specimens testing positive for influenza is not reported because public health laboratories often receive samples that have already tested positive for influenza at a clinical laboratory and therefore percent positive would not be a valid indicator of influenza activity. Additional information is available at http://www.cdc.gov/flu/weekly/overview.htm.
Novel Influenza A Virus:
One human infection with a novel influenza A virus was reported during week 51. The person was infected with an avian lineage influenza A (H7N2) virus. This person reported close, prolonged unprotected exposure to the respiratory secretions of infected, sick cats at an affected New York City animal shelter. This is the first influenza A (H7N2) virus infection in humans identified in the United States since 2003 and the first known human infection with an influenza A virus likely acquired through exposure to an ill cat. The patient was mildly ill, was not hospitalized, and has recovered completely. No human-to-human transmission has been identified.
An outbreak of avian lineage influenza A (H7N2) virus infection among cats in an animal shelter in New York City was first reported to public health on December 14, 2016. The New York City Department of Health and Mental Hygiene (DOHMH) issued a press release on December 22 that can be found here: http://www1.nyc.gov/site/doh/about/press/pr2016/pr107-16.page .
Early identification and investigation of human infections with novel influenza A viruses are critical so that the risk of infection can be more fully understood and appropriate public health measures can be taken. Additional information on influenza in cats can be found here (https://www.cdc.gov/flu/fluincats/index.htm)
and a web spotlight on avian influenza A (H7N2) in cats in animal shelters in NY can be found here (https://www.cdc.gov/flu/spotlights/avian-influenza-cats.htm)
.
!–>
Influenza Virus Characterization:
CDC characterizes influenza viruses through one or more tests including genomic sequencing, hemagglutination inhibition (HI) and/or neutralization assays. These data are used to compare how similar currently circulating influenza viruses are to the reference viruses used for developing influenza vaccines, and to monitor for changes in circulating influenza viruses. Historically, HI data have been used most commonly to assess the similarity between reference viruses and circulating viruses to suggest how well the vaccine may work until such a time as vaccine effectiveness estimates are available.
For nearly all virus positive surveillance samples received at CDC, next-generation sequencing is performed to ascertain genomic data of circulating influenza viruses. Viruses can be classified into genetic groups/clades based on analysis of their HA gene segments using phylogenetics and key amino acid changes (Klimov Vaccine 2012).
A proportion of influenza A (H3N2) viruses don’t yield sufficient hemagglutination titers for antigenic characterization using the hemagglutination inhibition test. Therefore, CDC selects a subset of influenza A (H3N2) viruses to test using a focus reduction assay for supplementary antigenic characterization.
Genetic Characterization
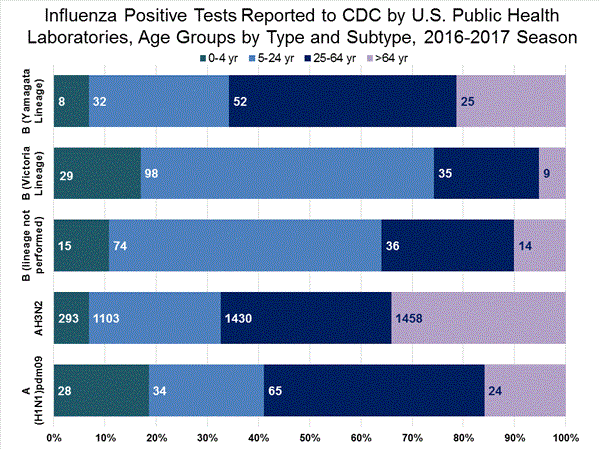
During the 2016-2017 season, 6,016 influenza positive specimens have been collected and reported by public health laboratories in the United States (figure, left). CDC genetically characterized 412 influenza viruses [44 influenza A (H1N1)pdm09, 299 influenza A (H3N2), and 69 influenza B viruses] collected by U.S. laboratories. The HA gene segment of all influenza A (H1N1)pdm09 viruses analyzed belonged to genetic group 6B.1. Influenza A (H3N2) virus HA gene segments analyzed belonged to genetic groups 3C.2a or 3C.3a. Genetic group 3C.2a includes a newly emerging subgroup known as 3C.2a1. The HA of influenza B/Victoria-lineage viruses all belonged to genetic group V1A. The HA of influenza B/Yamagata-lineage viruses analyzed all belonged to genetic group Y3.
The majority of U.S. viruses submitted for characterization come from state and local public health laboratories. Due to Right Size Roadmap considerations, specimen submission guidance issued to the laboratories request that, if available, 2 influenza A (H1N1), 2 A influenza (H3N2), and 2 influenza B viruses be submitted every other week. Because of this, the number of each virus type/subtype characterized should be approximately equal. In the figure below, the results of tests performed by public health labs are presented on the left and sequence results by genetic group of specimens submitted to CDC are presented on the right.
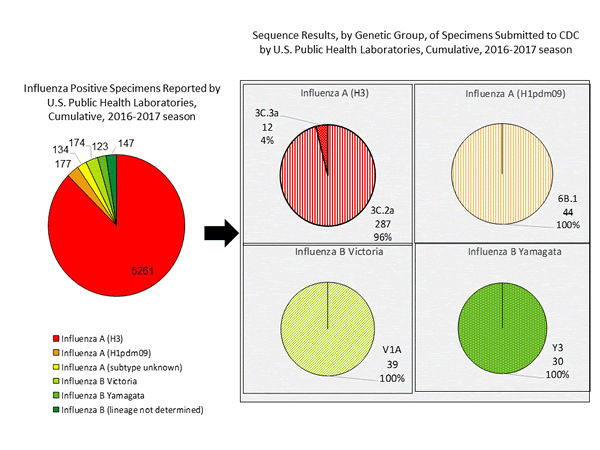
Antigenic Characterization
CDC has antigenically characterized 235 influenza viruses [37 influenza A (H1N1)pdm09, 139 influenza A (H3N2), and 59 influenza B viruses] collected by U.S. laboratories since October 1, 2016.
Influenza A Virus [176]
- A (H1N1)pdm09 [37]: All 37 (100%) influenza A (H1N1)pdm09 viruses were antigenically characterized using ferret post-infection antisera as A/California/7/2009-like, the influenza A (H1N1) component of the 2016-2017 Northern Hemisphere vaccine.
- A (H3N2) [139]: 132 of 139 (95.0%) influenza A (H3N2) viruses were antigenically characterized as A/Hong Kong/4801/2014-like, a virus that belongs in genetic group 3C.2a and is the influenza A (H3N2) component of the 2016-2017 Northern Hemisphere vaccine, by HI testing or neutralization testing. Among the viruses which reacted poorly with ferret antisera raised against A/Hong Kong/4801/2014-like viruses, 6 out of 7 (85.7%) are more closely related to A/Switzerland/9715293/2013, a virus belonging to genetic group 3C.3a.
Influenza B Virus [59]
- Victoria Lineage [31]: 28 of 31 (90.3%) B/Victoria-lineage viruses were antigenically characterized using ferret post-infection antisera as B/Brisbane/60/2008-like, which is included as an influenza B component of the 2016-2017 Northern Hemisphere trivalent and quadrivalent influenza vaccines.
- Yamagata Lineage [28]: All 28 (100%) B/Yamagata-lineage viruses were antigenically characterized using ferret post-infection antisera as B/Phuket/3073/2013-like, which is included as an influenza B component of the 2016-2017 Northern Hemisphere quadrivalent influenza vaccines.
<!–*CDC routinely uses hemagglutination inhibition (HI) assays to antigenically characterize influenza viruses year-round to compare how similar currently circulating influenza viruses are to those included in the influenza vaccine, and to monitor for changes in circulating influenza viruses. However, a portion of recent influenza A(H3N2) viruses do not grow to sufficient hemagglutination titers for antigenic characterization by HI. For many of these viruses, CDC is also performing genetic characterization to infer antigenic properties.–> <!–
2016-2017 Influenza Season – U.S. Influenza Vaccine Composition:
The World Health Organization (WHO) has recommended vaccine viruses for the 2016-2017 influenza season Northern Hemisphere vaccine composition, and the Food and Drug Administration’s Vaccines and Related Biological Products Advisory Committee (VRBPAC) has made the vaccine composition recommendation to be used in the United States. Both agencies recommend that trivalent vaccines contain an A/California/7/2009 (H1N1)pdm09-like virus, an A/Switzerland/9715293/2013 (H3N2)-like virus, and a B/Phuket/3073/2013-like (B/Yamagata lineage) virus. It is recommended that quadrivalent vaccines, which have two influenza B viruses, contain the viruses recommended for the trivalent vaccines, as well as a B/Brisbane/60/2008-like (B/Victoria lineage) virus. This represents a change in the influenza A (H3) and influenza B (Yamagata lineage) components compared with the composition of the 2016-2017 influenza vaccine. These vaccine recommendations were based on several factors, including global influenza virologic and epidemiologic surveillance, genetic characterization, antigenic characterization, antiviral resistance, and the candidate vaccine viruses that are available for production.
–>
Antiviral Resistance:
Testing of influenza A (H1N1)pdm09, influenza A (H3N2), and influenza B virus isolates for resistance to neuraminidase inhibitors (oseltamivir, zanamivir, and peramivir) is performed at CDC using a functional assay. Additional influenza A (H1N1)pdm09 and influenza A (H3N2) clinical samples are tested for mutations of the virus known to confer oseltamivir resistance. The data summarized below combine the results of both testing methods. These samples are routinely obtained for surveillance purposes rather than for diagnostic testing of patients suspected to be infected with antiviral-resistant virus.
High levels of resistance to the adamantanes (amantadine and rimantadine) persist among influenza A (H1N1)pdm09 and influenza A (H3N2) viruses (the adamantanes are not effective against influenza B viruses). Therefore, data from adamantane resistance testing are not presented below.
|
Oseltamivir |
Zanamivir |
Peramivir |
||||
|---|---|---|---|---|---|---|
|
Virus Samples tested (n) |
Resistant Viruses, Number (%) |
Virus Samples tested (n) |
Resistant Viruses, Number (%) |
Virus Samples tested (n) |
Resistant Viruses, Number (%) |
|
| Influenza A (H1N1)pdm09 |
51 |
0 (0.0) |
51 |
0 (0.0) |
51 |
0 (0.0) |
| Influenza A (H3N2) |
317 |
0 (0.0) |
317 |
0 (0.0) |
251 |
0 (0.0) |
| Influenza B |
81 |
0 (0.0) |
81 |
0 (0.0) |
81 |
0 (0.0) |
The majority of recently circulating influenza viruses are susceptible to the neuraminidase inhibitor antiviral medications, oseltamivir, zanamivir, and peramivir; however, rare sporadic instances of oseltamivir-resistant and peramivir-resistant influenza A (H1N1)pdm09 viruses and oseltamivir-resistant influenza A (H3N2) viruses have been detected worldwide. Antiviral treatment as early as possible is recommended for patients with confirmed or suspected influenza who have severe, complicated, or progressive illness; who require hospitalization; or who are at high risk for serious influenza-related complications. Additional information on recommendations for treatment and chemoprophylaxis of influenza virus infection with antiviral agents is available at http://www.cdc.gov/flu/antivirals/index.htm.
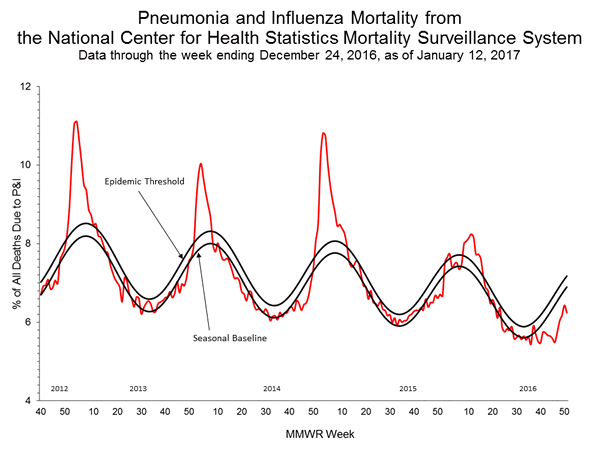
Pneumonia and Influenza (P&I) Mortality Surveillance:
Based on National Center for Health Statistics (NCHS) mortality surveillance data available on January 12, 2017, 6.2% of the deaths occurring during the week ending December 24, 2016 (week 51) were due to P&I. This percentage is below the epidemic threshold of 7.2% for week 51.
Background: Weekly mortality surveillance data includes a combination of machine coded and manually coded causes of death collected from death certificates. There is a backlog of data requiring manual coding within NCHS mortality surveillance data. The percentages of deaths due to P&I are higher among manually coded records than more rapidly available machine coded records and may result in initially reported P&I percentages that are lower than percentages calculated from final data. Efforts continue to reduce and monitor the number of records awaiting manual coding.
Beginning in the week ending October 8, 2016 (week 40), CDC retired the 122 Cities Mortality Reporting System and uses only the NCHS Mortality Surveillance System.
Region and state-specific data are available at http://gis.cdc.gov/grasp/fluview/mortality.html.
Influenza-Associated Pediatric Mortality:
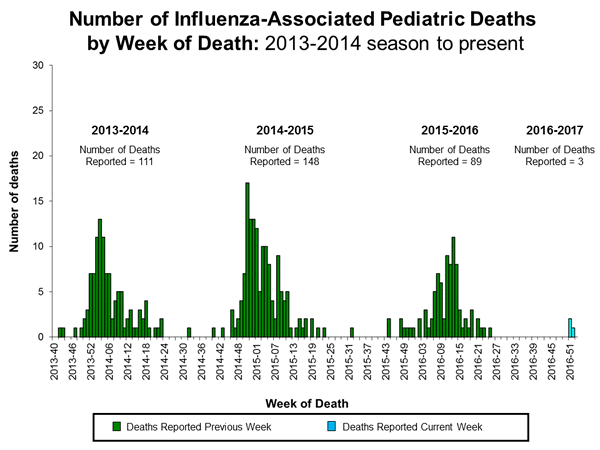
Three influenza-associated pediatric deaths were reported to CDC during week 1. One death was associated with an influenza A (H3) virus and occurred during week 51 (the week ending December 24, 2016). One death was associated with an influenza A virus for which no subtyping was performed and occurred during week 52 (the week ending December 31, 2016). One death was associated with an influenza B virus and occurred during week 51.
A total of three influenza-associated pediatric deaths have been reported for the 2016-2017 season.
Additional data can be found at: http://gis.cdc.gov/GRASP/Fluview/PedFluDeath.html.
Influenza-Associated Hospitalizations:
The Influenza Hospitalization Surveillance Network (FluSurv-NET) conducts population-based surveillance for laboratory-confirmed influenza-related hospitalizations in children younger than 18 years of age (since the 2003-2004 influenza season) and adults (since the 2005-2006 influenza season).
The FluSurv-NET covers more than 70 counties in the 10 Emerging Infections Program (EIP) states (CA, CO, CT, GA, MD, MN, NM, NY, OR, and TN) and additional Influenza Hospitalization Surveillance Project (IHSP) states. The IHSP began during the 2009-2010 season to enhance surveillance during the 2009 H1N1 pandemic. IHSP sites included IA, ID, MI, OK and SD during the 2009-2010 season; ID, MI, OH, OK, RI, and UT during the 2010-2011 season; MI, OH, RI, and UT during the 2011-2012 season; IA, MI, OH, RI, and UT during the 2012-2013 season; and MI, OH, and UT during the 2013-2014, 2014-2015, 2015-2016, and 2016-2017 seasons.
Data gathered are used to estimate age-specific hospitalization rates on a weekly basis, and describe characteristics of persons hospitalized with severe influenza illness. The rates provided are likely to be an underestimate as influenza-related hospitalizations can be missed, either because testing is not performed, or because cases may be attributed to other causes of pneumonia or other common influenza-related complications.
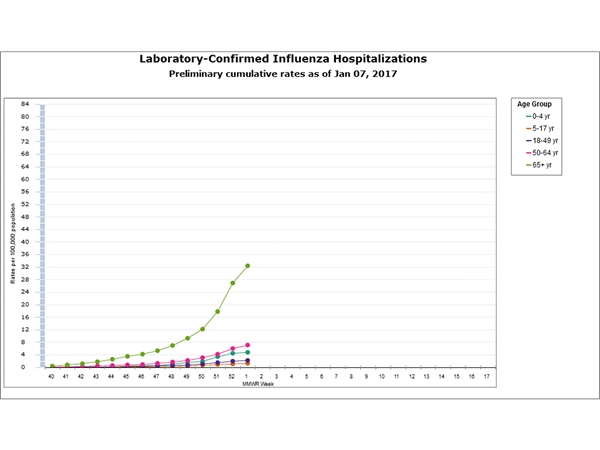
Between October 1, 2016 and January 7, 2017, 1,992 laboratory-confirmed influenza-associated hospitalizations were reported. The overall hospitalization rate was 7.1 per 100,000 population. The highest rate of hospitalization was among adults aged ≥65 years (32.4 per 100,000 population), followed by adults aged 50-64 (7.1 per 100,000 population) and children aged 0-4 years (4.8 per 100,000 population). Among 1,992 hospitalizations, 1,774 (89.1%) were associated with influenza A virus, 184 (9.2%) with influenza B virus, 11 (0.6%) with influenza A virus and influenza B virus co-infection, and 23 (1.2%) with influenza virus for which the type was not determined. Among those with influenza A subtype information, 493 (98.4%) were A(H3N2) and 8 (1.6%) were A(H1N1)pdm09 virus.
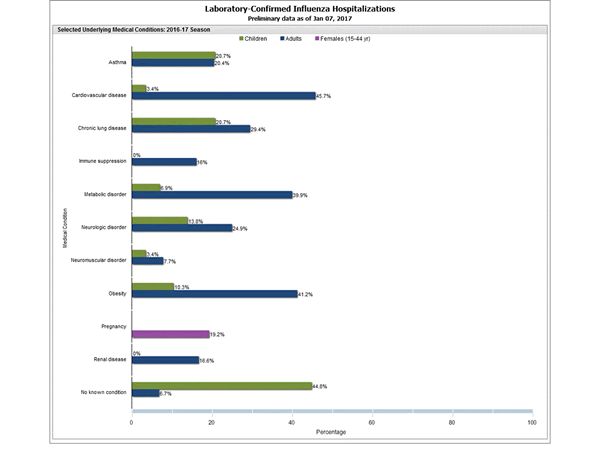
Clinical findings are preliminary and based on 336 (16.7%) cases with complete medical chart abstraction. Among 307 hospitalized adults with complete medical chart abstraction, 289 (94.1%) had at least one reported underlying medical condition; the most commonly reported were cardiovascular disease, obesity, and metabolic disorders. Among 29 hospitalized children with complete medical chart abstraction, 16 (55.2%) had at least one underlying medical condition; the most commonly reported were asthma and chronic lung disease. Among the 24 hospitalized women of childbearing age (15-44 years), 5 (15.8%) were pregnant.
Additional FluSurv-NET data can be found at: http://gis.cdc.gov/GRASP/Fluview/FluHospRates.html and http://gis.cdc.gov/grasp/fluview/FluHospChars.html.
Data from the Influenza Hospitalization Surveillance Network (FluSurv-NET), a population-based surveillance for influenza related hospitalizations in children and adults in 13 U.S. states. Cumulative incidence rates are calculated using the National Center for Health Statistics’ (NCHS) population estimates for the counties included in the surveillance catchment area.
FluSurv-NET data are preliminary and displayed as they become available. Therefore, figures are based on varying denominators as some variables represent information that may require more time to be collected. Data are refreshed and updated weekly. Asthma includes a medical diagnosis of asthma or reactive airway disease; Cardiovascular diseases include conditions such as coronary heart disease, cardiac valve disorders, congestive heart failure, and pulmonary hypertension; does not include isolated hypertension; Chronic lung diseases include conditions such as chronic obstructive pulmonary disease, bronchiolitis obliterans, chronic aspiration pneumonia, and interstitial lung disease; Immune suppression includes conditions such as immunoglobulin deficiency, leukemia, lymphoma, HIV/AIDS, and individuals taking immunosuppressive medications; Metabolic disorders include conditions such as diabetes mellitus; Neurologic diseases include conditions such as seizure disorders, cerebral palsy, and cognitive dysfunction; Neuromuscular diseases include conditions such as multiple sclerosis and muscular dystrophy; Obesity was assigned if indicated in patient’s medical chart or if body mass index (BMI) >30 kg/m2; Pregnancy percentage calculated using number of female cases aged between 15 and 44 years of age as the denominator; Renal diseases include conditions such as acute or chronic renal failure, nephrotic syndrome, glomerulonephritis, and impaired creatinine clearance; No known condition indicates that the case did not have any known high risk medical condition indicated in medical chart at the time of hospitalization.
Outpatient Illness Surveillance:
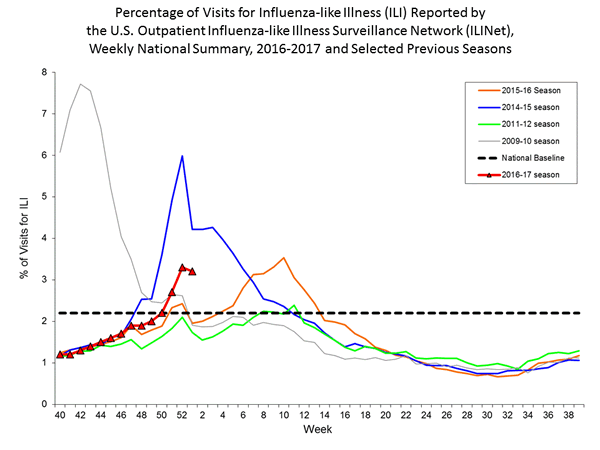
Nationwide during week 1, 3.2% of patient visits reported through the U.S. Outpatient Influenza-like Illness Surveillance Network (ILINet) were due to influenza-like illness (ILI). This percentage is above the national baseline of 2.2%.
(ILI is defined as fever (temperature of 100°F [37.8°C] or greater) and cough and/or sore throat.)
The increase in the percentage of patient visits for ILI may be influenced in part by a reduction in routine healthcare visits during the holidays, as has occurred in previous seasons.
Additional data are available at http://gis.cdc.gov/grasp/fluview/fluportaldashboard.html.
On a regional level, the percentage of outpatient visits for ILI ranged from 1.7% to 5.9% during week 1. All ten regions reported a proportion of outpatient visits for ILI at or above their region-specific baseline levels.
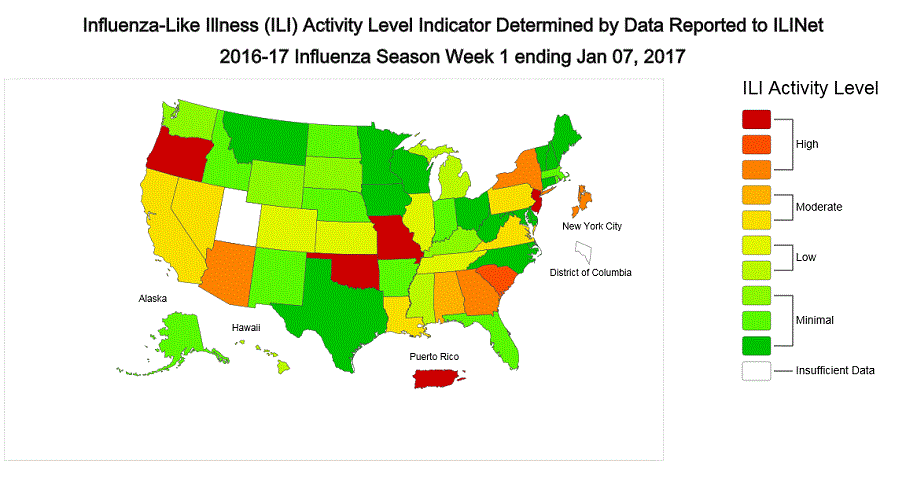
ILINet State Activity Indicator Map:
Data collected in ILINet are used to produce a measure of ILI activity* by state. Activity levels are based on the percent of outpatient visits in a state due to ILI and are compared to the average percent of ILI visits that occur during weeks with little or no influenza virus circulation. Activity levels range from minimal, which would correspond to ILI activity from outpatient clinics being below, or only slightly above, the average, to high, which would correspond to ILI activity from outpatient clinics being much higher than average.
During week 1, the following ILI activity levels were experienced:
- New York City, Puerto Rico, and eight states (Arizona, Georgia, Missouri, New Jersey, New York, Oklahoma, Oregon, and South Carolina) experienced high ILI activity.
- Six states (Alabama, California, Louisiana, Nevada, Pennsylvania, and Virginia) experienced moderate ILI activity.
- Seven states (Colorado, Hawaii, Illinois, Kansas, Michigan, Mississippi, and Tennessee) experienced low ILI activity.
- 28 states (Alaska, Arkansas, Connecticut, Delaware, Florida, Idaho, Indiana, Iowa, Kentucky, Maine, Maryland, Massachusetts, Minnesota, Montana, Nebraska, New Hampshire, New Mexico, North Carolina, North Dakota, Ohio, Rhode Island, South Dakota, Texas, Vermont, Washington, West Virginia, Wisconsin, and Wyoming) experienced minimal ILI activity.
- Data were insufficient to calculate an ILI activity level from the District of Columbia and one state (Utah).
<!–
View
Chart Data | View Full Screen
–>
*This map uses the proportion of outpatient visits to health care providers for ILI to measure the ILI activity level within a state. It does not, however, measure the extent of geographic spread of flu within a state. Therefore, outbreaks occurring in a single city could cause the state to display high activity levels.
Data collected in ILINet may disproportionally represent certain populations within a state, and therefore, may not accurately depict the full picture of influenza activity for the whole state.
Data displayed in this map are based on data collected in ILINet, whereas the State and Territorial flu activity map is based on reports from state and territorial epidemiologists. The data presented in this map is preliminary and may change as more data are received.
Differences in the data presented here by CDC and independently by some state health departments likely represent differing levels of data completeness with data presented by the state likely being the more complete.